
SPRUCE
The quality of results from any biophysical modeling project depends strongly on the ability to first prepare a system from an experimental data file, such as PDB or mmCIF. Unfortunately, these experiments often cannot resolve key pieces of the biological system. The missing details could be as innocent as the location of the protons, or as extreme as the missing entire mobile portions of a protein.
SPRUCE streamlines the preparation process by automatically breaking the system into individual biological components, adding any missing protons or residues, and it finishes by optimizing the hydrogen bond network for the entire system.
SPRUCE's structure preparation workflow performs tasks including the enumeration of biological units, alternate locations (if present), modeling missing residues and loops, and placing and optimizing hydrogens, accounting for the likely tautomer states of bound heterogens (ligands and cofactors).
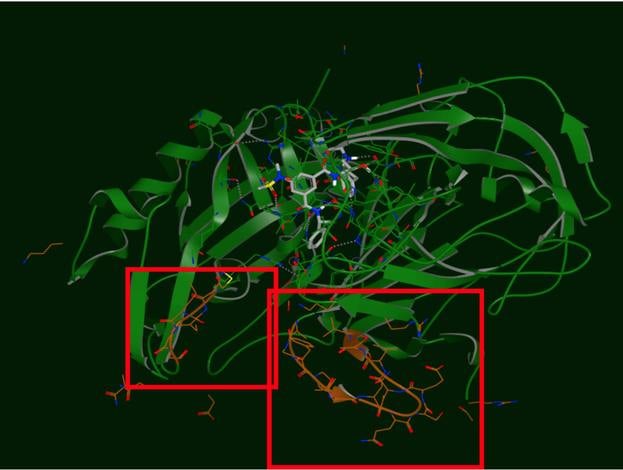
The produced output of SPRUCE is an OEDesignUnit. The OEDesignUnit has everything well componentized, making it easy to select which key pieces to include in the subsequent modeling tasks, and which to discard (e.g. excipients).
SPRUCE furthermore leverages the Iridium categorization [1] by providing the user with information about which structure is best to use for modeling. Additionally, SPRUCE highlights the parts of a structure that need special attention, if it is to be used in modeling.
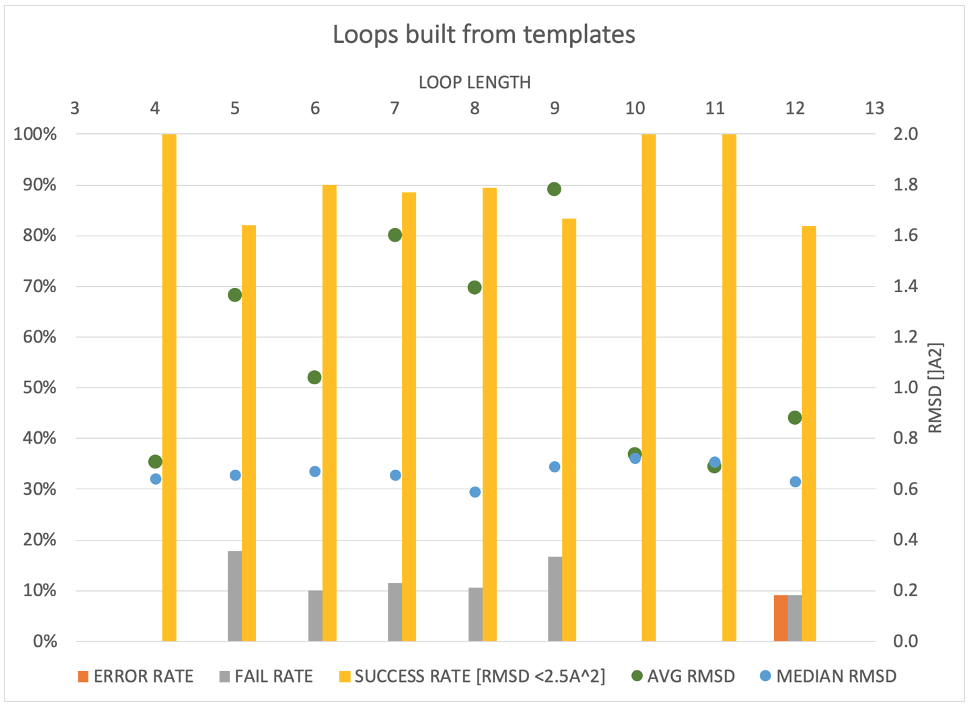
SPRUCE provides an expanding array of modeling tasks, like point-mutations, side-chain re-modeling and loop modeling using a template-based approach.
SPRUCE, furthermore, provides access to several superposition methods, based on sequence, secondary structures, or active site shape, each providing benefits depending on the similarity of the proteins being superposed.
Structure-based drug design requires careful preparation of experimental structures for downstream modeling applications. SPRUCE is a comprehensive, biomodeling preparation tool that reads experimentally solved (or modeled) protein and/or nucleic acid structures in several file formats and makes them modeling ready, for docking or molecule simulations.
To find out how SPRUCE can help with your protein modeling projects, contact us at info@eyesopen.com
DocumentationModeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can therefore be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid Comparison of Protein Binding Sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK General purpose optimization with MMFF94
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
Cheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all of the OpenEye applications and remaining toolkits are built. The Cheminformatics suite is a collection of seven individual yet interdependent toolkits that are described in the table below.
- FastROCS TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem TK name-to-structure, structure-to-name, foreign language translation
- Quacpac TK Tautomer enumeration and charge assignment
- MedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
References
- Essential considerations for using protein-ligand structures in drug discovery, G.L. Warren, T. D. Do, B. P. Kelly, A. Nicholls, S. D. Warren, Drug Discov. Today, 2012, 17, 1270-81
- Loopholes and missing links in protein modeling, A. Rossi, C. A. Weiglet, A. Nayeem, S. R. Krystek Jr., Prot. Sci., 2007, 1999-2012
RESOURCES
